This section of the AP Biology curriculum focuses on the process of transcription and the RNA processing mechanisms that stabilize a pre-mRNA molecule, remove the introns, and ready the molecule for export from the nucleus and translation into a protein molecule. We’ll start by taking a look at how RNA polymerase functions to create a primary RNA transcript from a DNA molecule. Then, we’ll see the three types of RNA (including messenger RNA, transfer RNA, and ribosomal RNA) that work together to translate information from the nucleic acid language to the amino acid language. Finally, we’ll see how alterations to the RNA processing mechanisms can create alternative splicing – different versions of a protein, based on the exons and introns that are kept in the finalized mRNA molecule.
The following video summarizes the most important aspects of this topic!
To watch more tutorial videos like this, please click here to see our full Youtube Channel!
ENDURING UNDERSTANDING
IST-1
Heritable information provides for continuity of life.
LEARNING OBJECTIVE
IST-1.N
Describe the mechanisms by which genetic information flows from DNA to RNA to protein.
ESSENTIAL KNOWLEDGE
IST-1.N.1
The sequence of the RNA bases, together with the structure of the RNA molecule, determines RNA function —
IST-1.N.2
Genetic information flows from a sequence of nucleotides in DNA to a sequence of bases in an mRNA molecule to a sequence of amino acids in a protein.
IST-1.N.3
RNA polymerases use a single template strand of DNA to direct the inclusion of bases in the newly formed RNA molecule. This process is known as transcription.
IST-1.N.4
The DNA strand acting as the template strand is also referred to as the noncoding strand, minus strand, or antisense strand. Selection of which DNA strand serves as the template strand depends on the gene being transcribed.
IST-1.N.5
The enzyme RNA polymerase synthesizes mRNA molecules in the 5’ to 3’ direction by reading the template DNA strand in the 3’ to 5’ direction.
IST-1.N.6
In eukaryotic cells, the mRNA transcript undergoes a series of enzyme-regulated modifications —
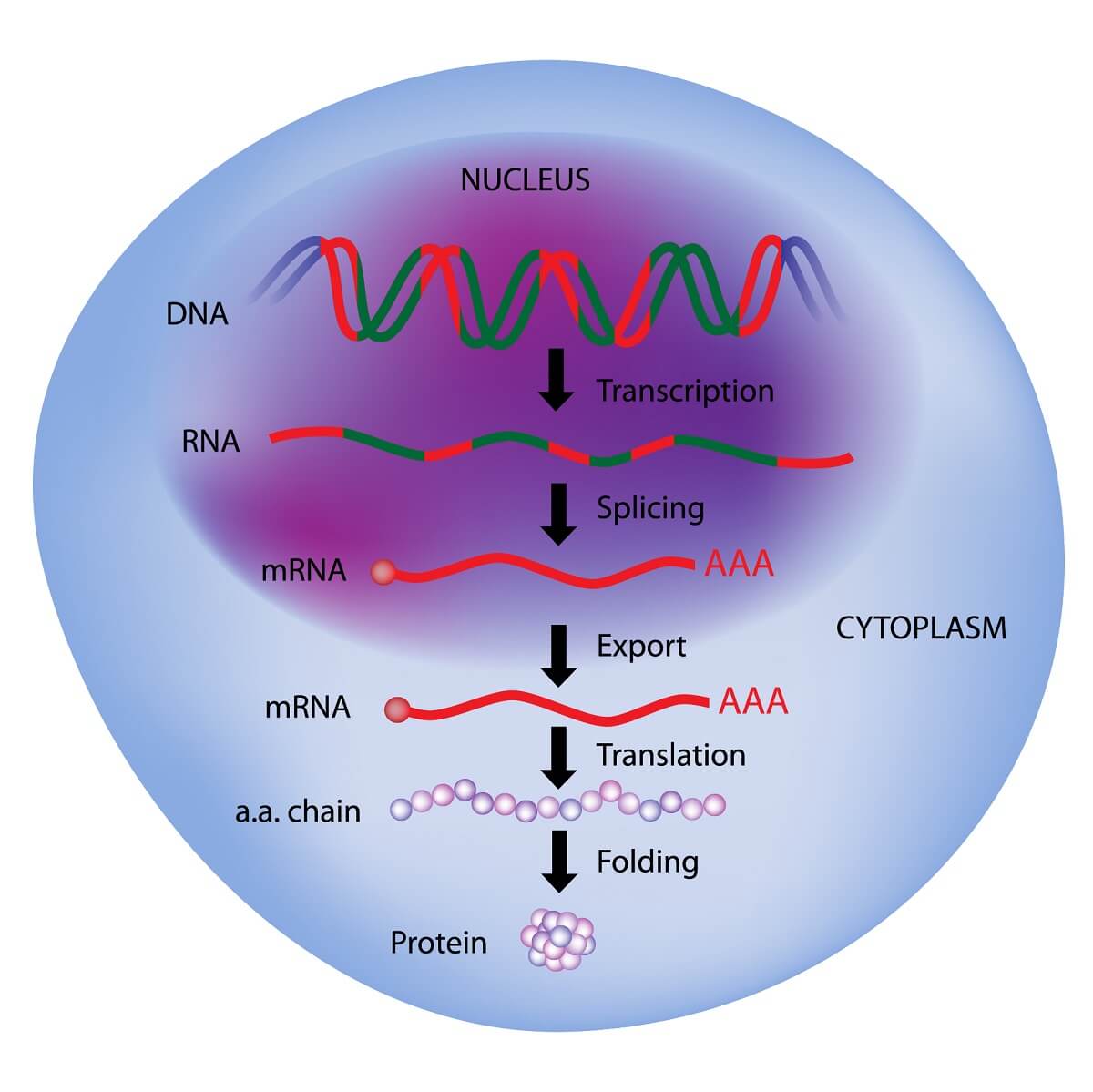
There are two main processes needed to extract the information from DNA and use it to create proteins. First, the information is transcribed in the nucleotide language from a DNA molecule into an RNA molecule. Transcription is like copying someone’s notes, it is the same information in the same language. Then, the RNA molecule must be translated – from the language of nucleotides into the language of amino acids – much like you would translate text from one language to another.
This section is going to examine the process of transcription, the enzymes needed for this process, the various forms of RNA involved, and the final step of RNA processing that is required before RNA can be translated into a protein. RNA transcription and processing will definitely be on the AP test. So, follow along as we cover everything you need to know about Transcription and RNA Processing!

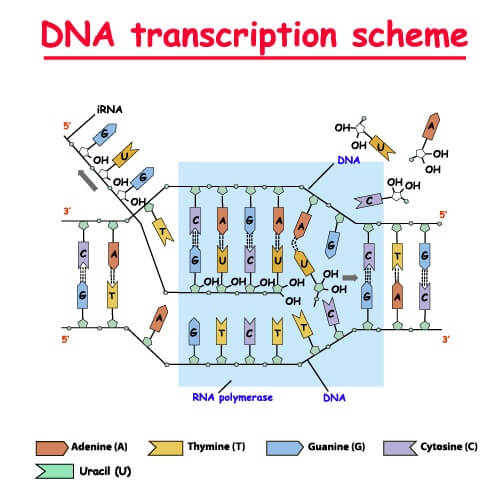
RNA Transcription is the process that copies a DNA sequence into a new RNA sequence, and it is very similar to DNA replication. Instead of DNA polymerase, this process uses RNA polymerase to separate the strands and synthesize a new RNA molecule from the DNA template. This RNA polymerase recognizes a promoter sequence in the DNA and starts the process of transcription from these points. This is known as initiation.
The RNA polymerase enzyme opens up a transcription bubble in the DNA strand, which allows free RNA nucleotides to hydrogen bond with the template strand. As each nucleotide of the template DNA strand hydrogen bonds to a complementary RNA nucleotide, RNA polymerase catalyzes the reaction to form the phosphodiester bond between the 3’ OH group of the growing RNA sequence and the 5’ phosphate group of the free nucleotide. This part of the RNA transcription process is known as elongation.
The final step of RNA transcription, known as termination, occurs when this RNA transcript separates from the transcription complex. This happens in a variety of complex ways in different organisms, but it is typically dependent on the sequence of nucleotides. Sometimes the sequence itself forms a tertiary structure called a hairpin that helps separate the RNA from the DNA, whereas other times a secondary protein recognizes a specific sequence in the RNA and physically separates the RNA from the template strand. This leaves a primary RNA transcript.
Keep in mind that this primary RNA transcript is an exact copy of the DNA strand that contains the code for a protein (called the “sense strand”). To make this exact copy, the DNA Template Strand is actually the antisense strand, since RNA polymerase can only operate by matching complementary nucleotides to the template strand.
One final thing to note is that the sense strand and the antisense strand designations rely entirely on the gene being transcribed. While the RNA transcript is always created in the 5’ to 3’ direction, different genes can use different strands of the DNA to serve as the template. RNA polymerase will simply recognize the promoter regions and move in the appropriate direction for each gene.
Think about this… each gene is kind of like a movie. When the movie is filmed, there are many scenes and takes that must be deleted from the final film. These are like the introns in a gene. They must be cut out of the primary RNA transcript and the exons must be spliced back together to make a functional RNA molecule. We’ll see this process in the second half of this video. For now, let’s consider the different types of RNA that a primary transcript can become.
The central dogma of biology states that DNA is transcribed into RNA and RNA is translated into protein. What this statement fails to mention is that there are many different kinds of RNA involved in the translation of the nucleic acid language to the amino acid language. Keep in mind that there are literally dozens of different kinds of RNA and RNA-based enzymes that serve roles in gene regulation and other cellular processes. But, there are three forms of RNA that are the most ubiquitous and important forms of RNA in most organisms.
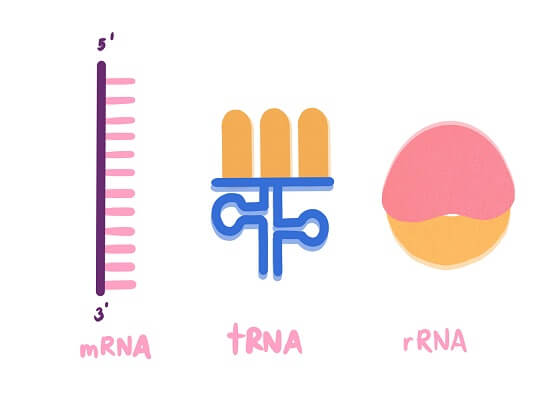
Messenger RNA (mRNA) is the form of RNA that actually stores the nucleotide sequence that can be translated into a protein sequence. It carries the genetic message from the DNA in the nucleus to the ribosomes outside of the nucleus to be translated. But, mRNA is just one piece of the puzzle.
The cell also needs ribosomal RNA (rRNA) and transfer RNA (tRNA) in order to translate this nucleic acid sequence into a sequence of amino acids. Ribosomal RNA forms a quaternary structure with several proteins to create a functional ribosome, as you can see here with the ribosomal RNA shown in light orange. Ribosomes have a large subunit and a small subunit, which come together around the mRNA molecule. The ribosomal RNA and proteins have specific areas that bind the mRNA molecule and areas that will catalyze the formation of a new protein chain. This is where transfer RNAs come into play.
Each tRNA carries a specific amino acid and has a specific anticodon sequence exposed to the mRNA. If all three of these anticodon nucleotides are perfect complements of the codon sequence, the tRNA molecule will add its amino acid to the growing polypeptide chain. Then, the empty tRNA will be ejected from the ribosome and recycled. As this happens, the ribosome moves along the mRNA molecule to the next codon, until it reaches a “STOP” codon and the sequence is finished. We’ll cover this process in more detail in section 6.4.
You should also note that each of these forms of RNA is stored in the genetic code. Each one is created by the process of transcription. However, the primary RNA transcript must undergo processing and specific modification before it can become mRNA, tRNA, or rRNA.
As noted in previous videos, the DNA code contains a lot more than just the information needed to create protein. For example, some estimates of the human genome suggest that only 8-15% of the genome actually contains sequences that become proteins (known as exons). The other 85% (or more) consist of regulatory elements (covered in section 6.5), telomeres, pseudogenes, repeat sequences, and introns. Some of these structures serve purposes, while others seem to be leftovers from the complexities of evolution. While most of these non-coding sequences lie between genes, the introns are non-coding sequences that lie within genes.
The process of RNA transcription creates a primary RNA transcript that still contains the introns. Since these introns contain non-coding DNA, leaving them in the mRNA molecule would create a nonsensical protein. Therefore, they must be removed through the process of splicing. The process of splicing starts with the removal of introns. The remaining exons are reassembled and bonded back together to create the pre-mRNA transcript. This sequence typically only contains coding regions, all arranged in the proper order to create a protein. This pre-mRNA needs a few things to become a mature mRNA molecule.
Two minor additions are made to this mRNA transcript before it is exported from the nucleus to become a protein. First, a molecule of GTP is added to the 5’ side of the RNA molecule. This stabilizes the molecule and blocks any random chemical reactions from taking place. On the 3’ side of the molecule, a long string of adenine nucleotides is added. Like the GTP molecule, this “poly-A tail” stabilizes the other side of the pre-mRNA. To do this, a number of proteins recognize specific sequences within the mRNA. These proteins recruit poly(A) polymerase which completes the process of polyadenylation.
Thus, with the introns spliced out, a GTP head, and a poly-A tail, the mRNA molecule is ready to leave the nucleus and be translated. Keep in mind that other forms of RNA, including tRNA and rRNA, also have specific post-transcription processing that folds them into the right shape and adds various elements.
The human genome encodes roughly 21,000 protein-coding genes. However, this is only a tiny fraction of the total variation created into actual proteins. This extra variation is created through the process of alternative splicing.
In regular splicing and processing, all of the exons for each gene get cut out and spliced back together to create the final mRNA transcript. To understand why this process can easily change, let’s take a closer look at how RNA processing actually works.
In most multicellular animals, the process of slicing is carried out by a complex enzyme structure known as a spliceosome. A spliceosome consists of 5 small-nuclear-RNA (snRNA) molecules and their associated proteins bound together into a complex. The spliceosome essentially binds to each intron, connecting the exons and removing the introns. If the spliceosome perfectly cuts out each intron and leaves each exon, one specific version of the protein is created. However, this process is loaded with complex variations.
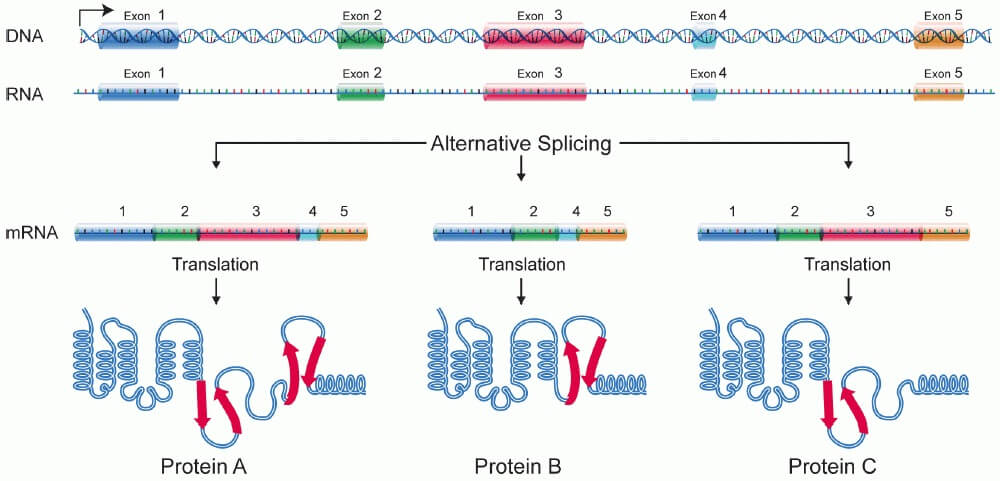
Each small-nuclear-RNA is genetically inherited with complex mutations. Plus, the proteins that create the spliceosome complex are also subject to variation. This can create spliceosomes that cut out certain exons entirely, leading to different isoforms of the same protein. These isoforms can also have extended exons or included introns. Some isoforms can function differently in different tissue types and environments, based on the spliceosome each cell creates – leading to differential cell functions. The many versions of a protein created by the inclusion of different exons and introns are made possible by alternative splicing.
More and more studies are revealing that alternatively spliced proteins account for nearly two-thirds of the genes in mice and humans. Each alternatively spliced protein can have between 2-25+ different variants – allowing certain proteins to function much differently in different cell types.